PhD, MSc Projects (ongoing and future)
Areas of scientific activity:
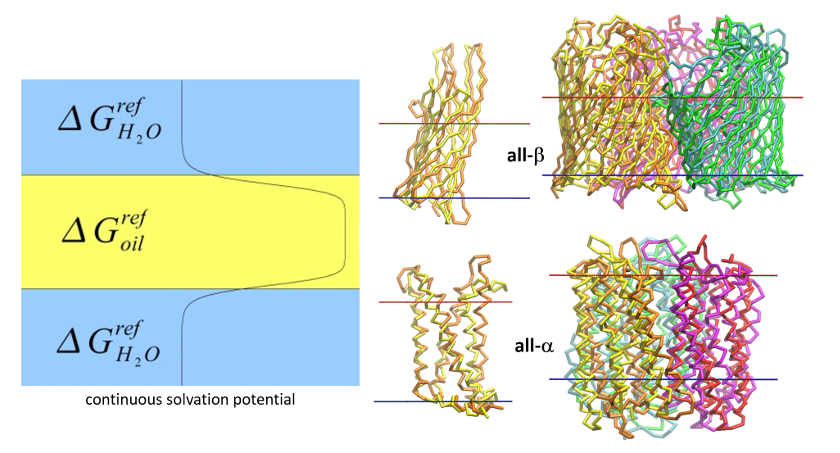
Modeling of structure and dynamics of membrane proteins and their complexes with ligands and other proteins. Drug design. Development of new methods of coarse-grain dynamics. Studies of interactions of proteins with graphene, carbon nanotubes and other electrode materials for biosensor applications.
Exemplary titles of expected Master/Doctoral theses:
- Modeling of action of agonist/antagonist sensor and molecular switches of formyl / opioid / cannabinoid / chemokine / etc. receptors from GPCR family using theoretical methods.
- Studies of formation and dynamics of oligomers of membrane proteins.
- Design of drugs preventing aggregation of beta-amyloid / activating or deactivating GPCRs / etc.
- Interactions of modified graphene or other electrode nanomaterial with proteins.
Tematyki badawcze:
Modelowanie struktury i dynamiki białek błonowych oraz ich kompleksów z ligandami oraz z innymi białkami. Projektowanie leków. Rozwój nowych metod symulacji gruboziarnistych. Badanie oddziaływań białek z grafenem, nanorurkami i innymi materiałami elektrodowymi do zastosowania w bioczujnikach.
Przykładowe przewidywane tematy prac magisterskich/doktorskich:
- Modelowanie działania sensora agonista/antagonista oraz przełączników molekularnych receptorów formylowych / opioidowych / kanabinoidowych / chemokinowych i innych z rodziny GPCR metodami teoretycznymi.
- Badanie powstawania i dynamiki oligomerów białek błonowych.
- Projektowanie leków przeciwdziałających agregacji beta-amyloidu / aktywujących lub deaktywujących receptory GPCR i inne.
- Oddziaływanie modyfikowanego grafenu lub innego nanomateriału elektrody z białkami.
The applicants interested in working in our group are welcomed to contact us
Minimal requirements for candidates:
- Practical knowledge of at least one protein visualizing/modeling program (e.g. YASARA, VMD, PyMol, MolMol, SPDBviewer)
- Knowledge of types of protein structures
- Practical usage of biological web servers (e.g. UniProt, ExPASy, BLAST, Protein Data Bank)
- Basic programming skills (e.g. Python, Perl, C, C++, Fortran)
- Good knowledge of English
- July 2023.GS-SMD web server
for SMD simulations of γ-secretase complex.
Publication in Nucleic Acids Research 2023 Web server issue.
- August 2022.COGRIMEN
- June 2021.GPCRsignalOur new service GPCRsignal was recently published in NAR 2021, W1.