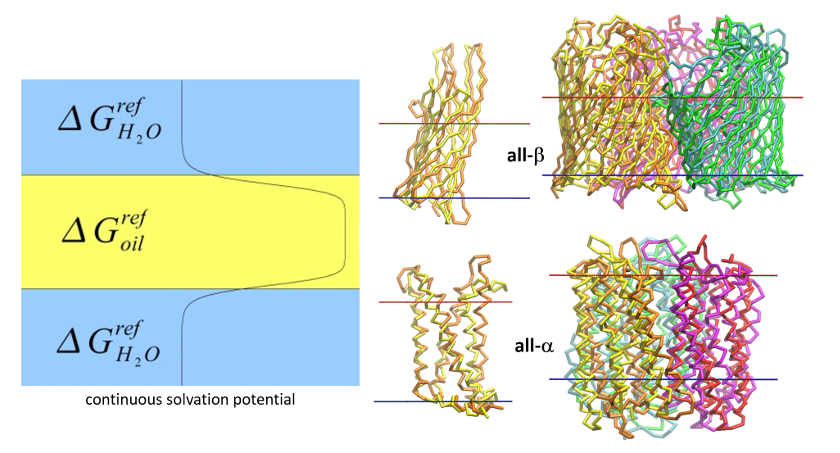
Scientific interests
- Drug design
- Modeling of structure and dynamics of membrane proteins
- Action of molecular switches on activation pathway of GPCRs, agonist/antagonist sensors
- Development of new methods for coarse-grain dynamics
- Oligomerization processes of proteins and peptides
- Interactions of proteins with graphene, carbon nanotubes and other electrode materials
- Simulated mechanical unfolding of proteins
Major investigated proteins
- G-protein-coupled receptors - GPCRs (rhodopsin, adrenergic / opioid / cannabinoid / chemokine / … receptors)
- Bacteriorhodopsin, halorhodopsin
- Membrane proteases including the γ-secretase complex (Alzheimer disease)
- Glucose oxidase (for biosensors)
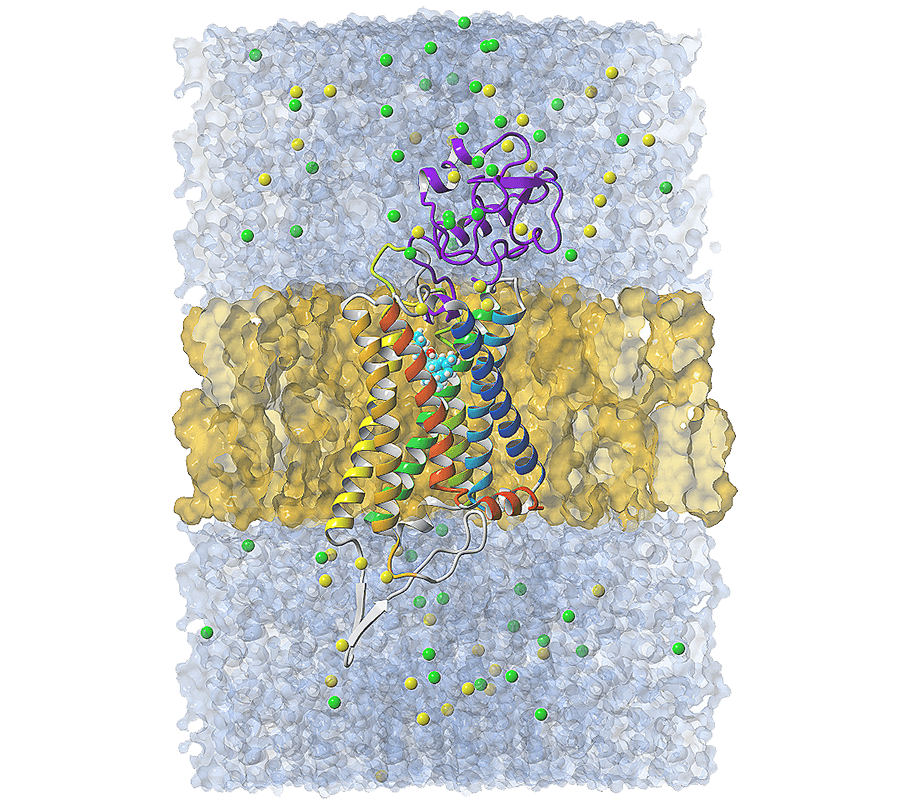
Graphics created by Jakub Jakowiecki
G protein-coupled receptor (GPCR) with a membrane, water, and ions, in a periodic box.
THE LATEST NEWS
- July 2023.GS-SMD web server
for SMD simulations of γ-secretase complex.
Publication in Nucleic Acids Research 2023 Web server issue.
- August 2022.COGRIMEN
- June 2021.GPCRsignalOur new service GPCRsignal was recently published in NAR 2021, W1.