Group Members > Jakub Jakowiecki
Jakub Jakowiecki, PhD

e-mail address: jjakowiecki|chem.uw.edu.pl
phone# +48 22 552 64 06 (Chemistry),
Faculty of Chemistry
& Biological and Chemical Research Centre,
University of Warsaw, Poland.
SCOPUC
ORCID
SCHOLAR
REPOZYTORIUM UW
Linkedin
born - Feb 6, 1983
married ????
Education:
- 2012-2023 University of Warsaw, Faculty of Chemistry, doctoral studies, specialization: computational chemistry (computer modeling of biomolecules), degree: doctor of natural sciences in the discipline of chemical sciences (awarded in 2023). Research for PhD thesis was performed in Biomodeling group, supervised by prof. dr. hab. Sławomir Filipek; Title of PhD thesis: Investigation of ligand binding to the CB1 cannabinoid receptor and other lipid GPCRs using molecular modeling methods
- 2002-2008 M.Sc. Study, University of Warsaw, Inter faculty Individual Studies in Mathematics and Natural Sciences on the University of Warsaw (MISMaP), major faculty: chemistry; Master's degree in organic chemistry (awarded in 2008). Research for master thesis was carried out in prof. dr. hab. Mieczysław Mąkosza group in Institute of Organic Chemistry, Polish Academy of Sciences; Title of master thesis: Synthesis of α-Trifluoromethyl-β-lactams and Esters of β Aminoacids via 1,3-Dipolar Cycloaddition of Nitrones to Penta- and Hexafluoropropene
- 1998-2002 Prince Józef Poniatowski High School (VLO) in Warsaw, profile: biological and chemical (graduated in 2002)
Employment:
- ????
Professional memberships:
- ????
Publication achievements:
- Orzeł U, Pasznik P, Miszta P, Lorkowski M, Niewieczerzał S, Jakowiecki J, Filipek S “GS-SMD server for steered molecular dynamics of peptide substrates in the active site of the γ-secretase complex” Nucleic Acids Res. 2023 May 19; doi:10.1093/nar/gkad409
- Jakowiecki J, Orzeł U, Gliździnska A, Możajew M, Filipek S. “Specificities of Protein Homology Modeling for Allosteric Drug Design” Methods Mol. Biol. 2023; 2627:339-348. doi:10.1007/978-1-0716-2974-1_19.
- Miszta, P; Pasznik, P; Niewieczerzal, S; Jakowiecki, J; Filipek, S; “GPCRsignal: webserver for analysis of the interface between G protein-coupled receptors and their effector proteins by dynamics and mutations” Nucleic Acids Res. 2021, 49, W247-W256, doi:10.1093/nar/gkab434
- Jakowiecki, J.; Abel, R.; Orzel, U.; Pasznik, P.; Preissner, R.; Filipek, S. Allosteric Modulation of the CB1 Cannabinoid Receptor by Cannabidiol-A Molecular Modeling Study of the N-Terminal Domain and the Allosteric-Orthosteric Coupling. Molecules 2021, 26, doi:10.3390/molecules26092456.
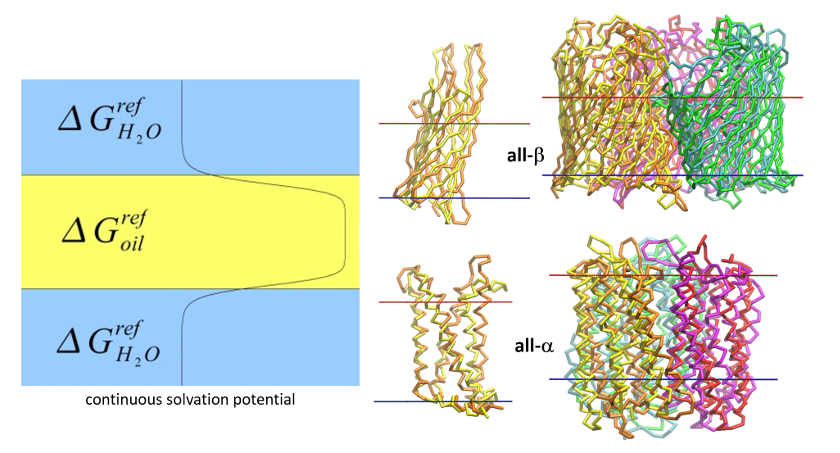
- Jakowiecki, J.; Orzel, U.; Chawananon, S.; Miszta, P.; Filipek, S. The Hydrophobic Ligands Entry and Exit from the GPCR Binding Site-SMD and SuMD Simulations. Molecules 2020, 25, doi:10.3390/molecules25081930.
- Jakowiecki J, Miszta P, Niewieczerzał S, Filipek S “Structural diversity in ligand recognition by GPCRs” (a chapter in a book: GPCR’s) 2020 doi:10.1016/B978-0-12-816228-6.00003-9
- Latek D, Trzaskowski B, Niewieczerzał S, Miszta P, Młynarczyk K, Dębiński A, Puławski W, Yuan S, Sztyler A, Orzeł U, Jakowiecki J, Filipek, S. “Modeling of Membrane Proteins: From Bioinformatics to Molecular Quantum Mechanics” (a chapter in a book: Computational Methods to Study the Structure and Dynamics of Biomolecules and Biomolecular Processes) 2019
- Miszta, P.; Pasznik, P.; Jakowiecki, J.; Sztyler, A.; Latek, D.; Filipek, S. GPCRM: a homology modeling web service with triple membrane-fitted quality assessment of GPCR models. Nucleic Acids Res. 2018, 46, W387-W395, doi:10.1093/nar/gky429.
- Jakowiecki J, Sztyler A, Filipek S, Li P, Raman K, Barathiraja N, Ramakrishna S, Eswara JR, Altaee A, Sharif AO, Ajayan PM and Renugopalakrishnan V. "Aquaporin–graphene interface: relevance to point-of-care device for renal cell carcinoma and desalination". 2018, Interface Focus 8(3): 20170066. doi:10.1098/rsfs.2017.0066
- Miszta P, Jakowiecki J, Rutkowska E, Turant M, Latek D, Filipek S. "Approaches for Differentiation and Interconverting GPCR Agonists and Antagonists" Methods Mol. Biol. 2018;1705:265-296. doi: 10.1007/978-1-4939-7465-8_12.
- Jarmuła A, Wilk P, Maj P, Ludwiczak J, Dowierciała A, Banaszak K, Rypniewski W, Cieśla J, Dąbrowska M, Frączyk T, Bronowska A K, Jakowiecki J, Filipek S, Rode W " Crystal structures of nematode (parasitic T. spiralis and free living C.elegans), compared to mammalian, thymidylate synthases (TS).Molecular docking and molecular dynamics simulations in search fornematode-specific inhibitors of TS" J. Mol. Graph. Model. (2017) 77, 33–50
- Rutkowska E, Miszta P, Młynarczyk K, Jakowiecki J, Pasznik P, Filipek S, Latek D "Application of a Membrane Protein Structure Prediction Web Service GPCRM to a Gastric Inhibitory Polypeptide Receptor Model" Lecture Notes in Computer Science Conference Paper (2017) . DOI: 10.1007/978-3-319-56154-7_15
- Jakowiecki J, Filipek S. "Hydrophobic ligands entry and exit pathways of CB1 cannabinoid receptor" J. Chem. Inf. Model. (2016), 56(12) · doi:10.1021/acs.jcim.6b00499
- Jakowiecki J, Loska R, Mąkosza M, "Synthesis of Trifluoromethyl--lactams and Esters of -Amino Acids via 1,3-Dipolar Cycloaddition of Nitrones to Fluoroalkenes". J. Org. Chem. (2008), 73(14), 5436-5441.
- Jankowiak A, Jakowiecki J, Kaszyński P, "Preparation of 4 Alkoxy-1-hydroxypyridine-2-thiones". Polish Journal of Chemistry, (2007), 81, 1869 1877.
Scientific interests:
- structure and dynamics of membrane proteins,
- G protein coupled receptors
- drug design,
- receptor activation processes,
- molecular dynamics simulations,
- artificial intelligence
Other interests:
- astronomy and astrophotography
- music
THE LATEST NEWS
- July 2023.GS-SMD web server
for SMD simulations of γ-secretase complex.
Publication in Nucleic Acids Research 2023 Web server issue.
- August 2022.COGRIMEN
- June 2021.GPCRsignalOur new service GPCRsignal was recently published in NAR 2021, W1.